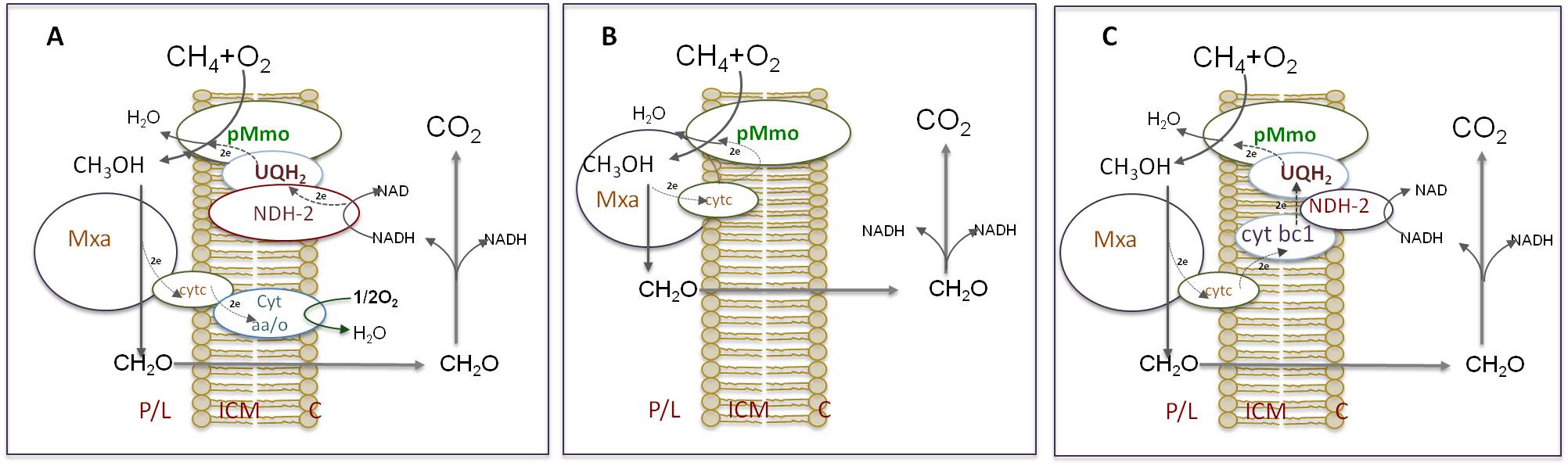
Genome scale FBA was used to test three possible modes of methane oxidation: a) redox–arm mode, the currently accepted model in which electrons driving methane oxidation come from NADH produced by formate or formaldehyde oxidation, while electrons produced from methanol oxidation are linked to a redox-arm and used for ATP production (adapted from Semrau et al., 2010); b) direct coupling mode, in which methanol oxidation supplies electrons for methane oxidation; and c) uphill electron transfer model, in which methanol oxidation partially supports methane oxidation (from Leak&Dalton, 1986). Proton translocation and ATP synthesis were omitted for simplicity. P – periplasm, L – ICM (lumen), C – cytoplasm.
The flux balance simulations suggest that only the transfer of electrons from methanol oxidation to methane oxidation steps can support measured growth and methane/oxygen consumption parameters, while the scenario employing NADH as a possible source of electrons for particulate methane monooxygenase cannot. Direct coupling between methane oxidation and methanol oxidation accounts for most of the membrane-associated methane monooxygenase activity. However the best fit to experimental results is achieved only after assuming that the efficiency of direct coupling depends on growth conditions and additional NADH input (about 0.1–0.2 mol of incremental NADH per one mol of methane oxidized). The additional input is proposed to cover loss of electrons through inefficiency and to sustain methane oxidation at perturbations or support uphill electron transfer.