The polymerase chain reaction (PCR)1 is a trick for producing relatively large amounts of a specific DNA or RNA sequence from only a few molecules of template. (Keep in mind that "relatively large amounts" typically means µg of the DNA or RNA.) Thus, PCR is said to "amplify" a particular sequence. PCR has a enormous number of practical applications. For example, PCR can be used for cloning specific genes, for sequencing genes, for studying gene expression, for mapping mutations, and for making mutations. Because of its broad impact on biology, Kary Mullis recieved a Nobel Prize in 1993 for his insight in developing this technique.
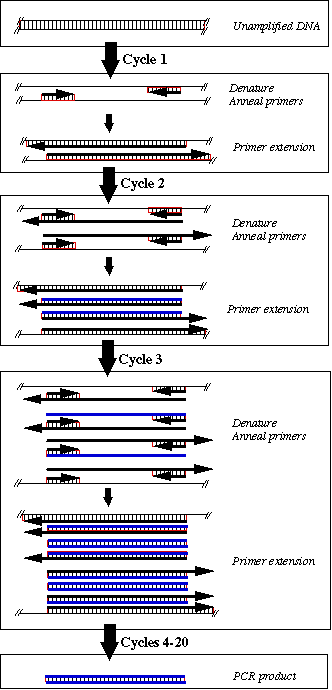
PCR amplification of DNA occurs by repeated cycles of three temperature dependent steps: (1) the double-stranded DNA (dsDNA) template is denatured; (2) oligonucleotide primers are annealed to the single-stranded DNA (ssDNA) template (one primer is designed to anneal to a specific region on the left side of one of the strands of DNA and the other primer is designed to anneal to a specific region on the right side of the complementary strand of DNA); and (3) DNA polymerase extends each primer in the 5' to 3' direction, duplicating the DNA fragment between the primers. With each cycle of denaturing, annealing, and synthesis the specific DNA fragment is amplified exponentially. The basic idea is shown in the following figure.

DNA synthesized during the first cycle has the 5' end of the primers and a variable 3' end. When these strands are denatured, the parental strand will rehybridize to the primer, so the product with a variable 3' end will continue to be synthesized during subsequent cycles of PCR. (Only one copy of each of the products with a variable 3' end will accumulate with each cycle.) The second cycle of denaturation, annealing, and primer extension produces discrete products with the 5' end of one primer and the 3' end of the other primer. Each strand of this discrete product is complementary to one of the two primers and thus acts as a template in subsequent cycles. Therefore, these products with defined 5' and 3' ends accumulate exponentially with each round of DNA amplification -- that is, for every n cycles of PCR there will be 2n-fold amplification of the specific DNA fragment.

The purity and yield of the PCR products depend on each of the reaction conditions: the temperature during each step of the cycle; the purity and sequence of the DNA template; the length and sequence of the oligonucleotide primers; the DNA polymerase used; the concentration of Mg++, the buffer, and other chemicals added to the reaction; and the number of cycles of PCR.
1The PCR process is patented by Hoffmann-La Roche Inc. Use of the PCR process requires a license. For an interesting discussion of the legal controversy over this patent, check out the article on the patent dispute from The Scientist.
lease send comments, suggestions, or questions to smaloy@sciences.sdsu.edu
Last modified July 15, 2002